A Beginner's Guide to Optuna
Many sophisticated algorithms, particularly deep learning-based ones, have various hyperparameters that control the running process. Those hyperparameters often have a significant impact on performance. Manually searching for a good combination of those hyperparameters is tedious, and even keeping track of all results becomes unwieldy as the number of hyperparameters grows beyond 10. Today's topic, Optuna, is such a tool that helps automate the search process and stores all results in a structured database. Even better, its web-based dashboard lets us explore these results intuitively with just a few clicks.
In this post, we'll first review the Optuna's basic workflow, revolving around several core concepts. Then we'll demonstrate its basic usage with a simple random search example. Advanced features are highlighted at the end, along with links to online resources for interested readers.
The Basic Workflow
Optuna is an open source hyperparameter optimization framework to automate hyperparameter search. See this video for a quick overview of Optuna's motivation and features. See their paper (Akiba et al., 2019) for the design philosophy. For interested readers, we recommend going through key features and recipes in the official tutorial.
To set up the hyperparameter optimization problem, we need an
evaluation metric \(c(\lambda)\) to measure the performance of a
hyperparameter combination \(\lambda \in \Lambda\). A hyperparameter optimization
algorithm explores a subset of \(\Lambda\) and tries to find a well-performed
\(\lambda\). In Optuna, this workflow is abstracted by two concepts, the study
and trials. A Study instance is responsible for optimizing a
user-defined objective function (i.e., the evaluation metric),
and cordinates multiple Trial objects to explore the hyperparameter
space \(\Lambda\). Each Trial is assigned a hyperparamter to be evaluated and
during the evaluation process some trials might be pruned to
accelerate the optimization process.
Sampling and Pruning are another two central concepts in
Optuna. Candidates of hyperparameter combinations are selected through
various sampling strategies, including uniform samplers for grid
search, random samplers for random search, samplers derived from
Bayesian optimization, and etc. Each candidate of hyperparameter
combinations is evaluated through a Trial object, and pruning
strategies might be applied to terminate underperforming trials early
to save computational resources.
Intuitively, Optuna's workflow can be summarized into following steps.
- Propose a hyperparameter combination. This is done by samplers.
- Evaluate the hyperaprameter combination. This is designated to
Trialinstances. - Record the trial result and propose new hyperparameter
combinations. In this step, the
Studyobject communicate with all trials and saves results in memory or a database. Moreover, various samplers used in Optuna can efficiently utilize history infomation to propose possible better hyperparameter candidates for future trials.
Within this framework, Optuna supports easy parallelization by running multiple trials simultaneously, efficient pruning by terminating trials early, and visualization by a separate dashboard module which interfaces with the database.
A Random Search Example
Below we demonstrate how to do random search with Optuna. For clarity, we focus on essential code snippets; complete scripts are available here. For more examples, please see the official repo. In particular, here is a simple example that optimizes a classifier configuration for Iris dataset using sklearn, and here is an official example that optimizes multi-layer perceptrons using PyTorch.
Set up the problem
Consider a simple nonlinear regression task which fits the sine function with fully-connect networks. We begin by a concise script to complete the training and testing. Once the original supervised learning problem is well-defined, we apply Optuna to tune its hyperparameters.
Original regression problem. Fit \(g(x)=\sin x\) with a fully-connected network \(f(x;\theta)\). The training dataset is generated by uniformly discretizing \([-4\pi, 4\pi]\) and has 800 data points. The test dataset is generated similarly but has 199 data points. Train a two-layer FC network and apply SGD for optimization.
# import necessary packages # define some global variables for convenience def get_dataloader(): """Return the train dataloader and test dataloader.""" def get_model_and_optimizer(config: dict): """Return the model and optimizer specified by `config`.""" def train_and_eval(model, optimizer, train_loader, test_loader, config): """Train the model with optimizer and report test error."""
Define the objective
The hyperparameter optimization problem. The hyperparameters to be tuned are 1) the batch size; 2) the hidden size; 3) the learning rate; 4) the momentum in SGD; 5) the number of training epochs. The objective of hyperparameter optimization is defined by averaging the test errors of 5 independent runs.
def objective(trial: optuna.trial.Trial): config = { "batch_size": trial.suggest_int("batch_size", 16, 128, step=16), "hidden_size": trial.suggest_int("hidden_size", 64, 512, step=64), "lr": trial.suggest_float("lr", 5e-5, 5e-3, log=True), "momentum": trial.suggest_float("momentum", 0.8, 0.99), "num_epochs": trial.suggest_int("num_epochs", 500, 5000, step=500), } train_loader, test_loader = get_dataloader() total_error = 0.0 for _ in range(NUM_REPEATS): model, optimizer = get_model_and_optimizer(config) test_error = train_and_eval(model, optimizer, train_loader, test_loader, config) total_error += test_error return total_error / NUM_REPEATS
This objective function accepts a Trial instance and uses optuna's
suggest API to define the search space of hyperparameters. During the
runtime, we need to specify a sampler to sample the hyperparameter
combination for this trial; see the doc for an overview of
samplers. Note that the value returned by suggest_int and
suggest_float is a single hyperparameter value sampled from a
distribution instead of a fixed set of values.
Create a study
We create a Study instance and equip it with the random sampler.
study = optuna.create_study(
sampler=optuna.samplers.RandomSampler(),
)
Then, we invoke the optimize method to run trials and evaluate
hyperparameter combinations sampled by the random sampler. For
demonstration purpose, we run 10 trials here. Besides the number of
trials, the optimize method also accept a timeout argument to limit
the elapsed time; see the doc for more details.
study.optimize(objective, n_trials=10)
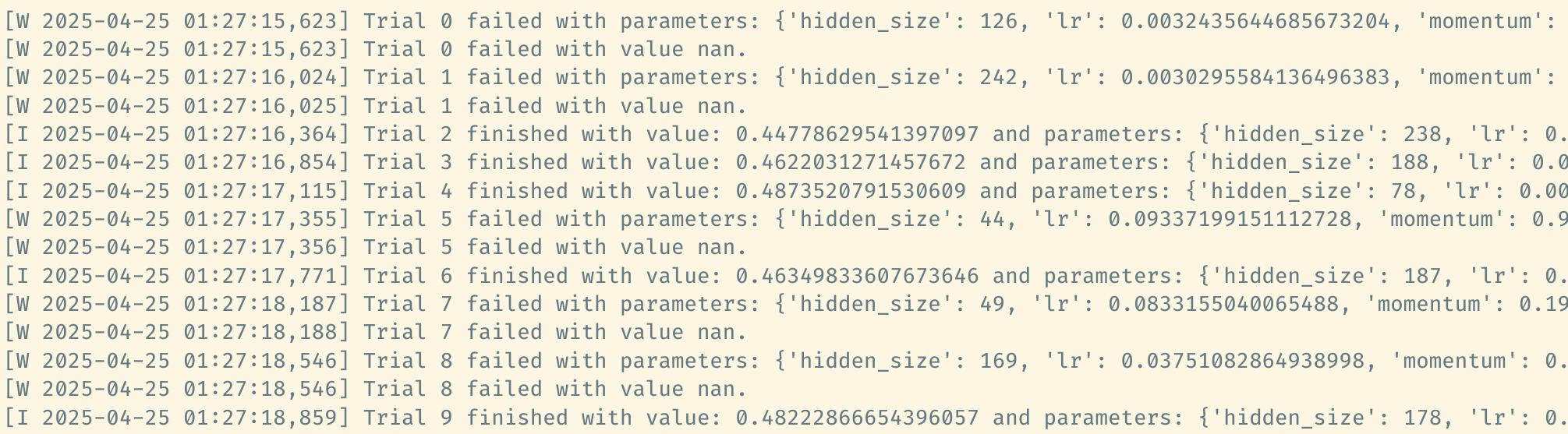
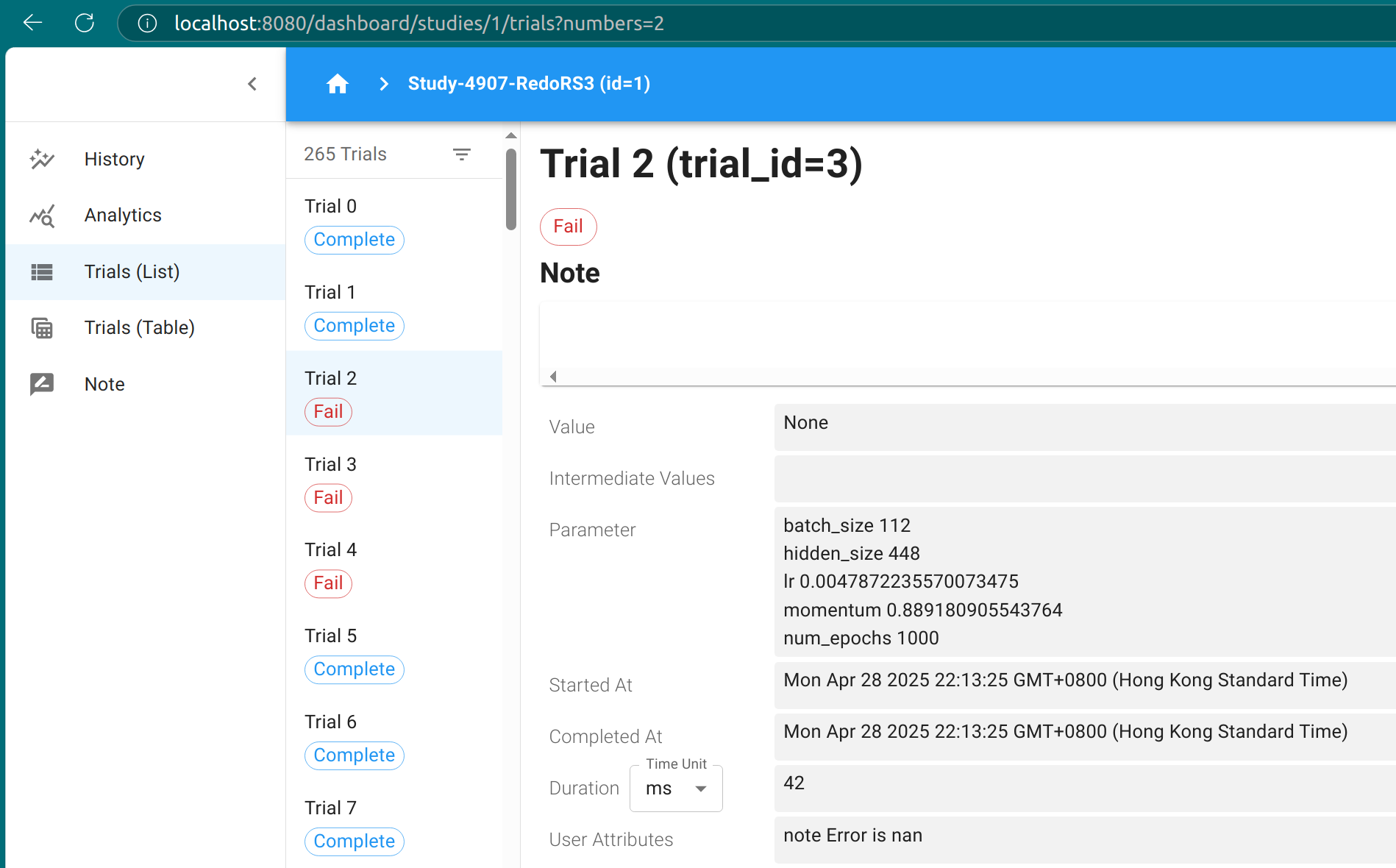
Inspect trial results
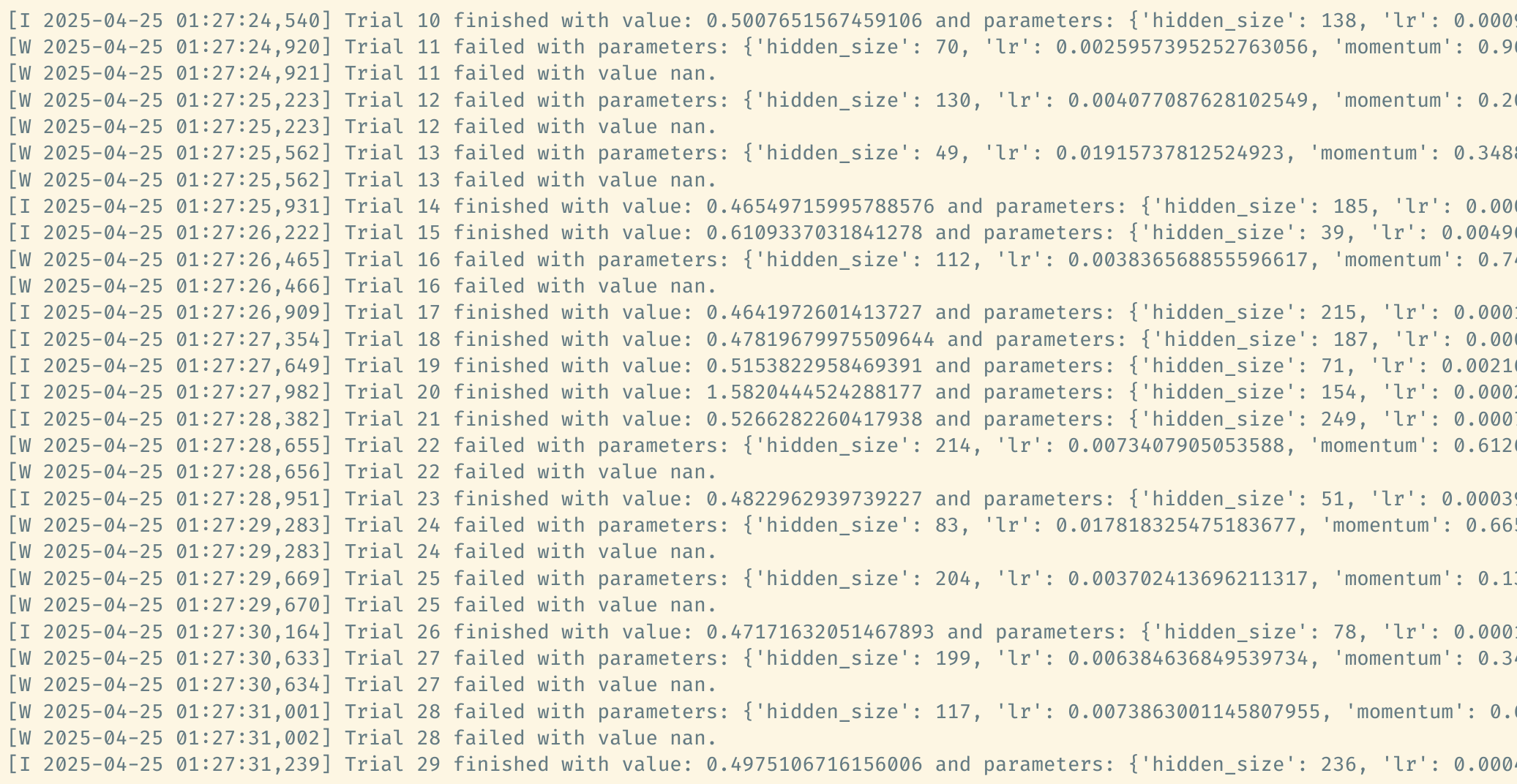
All evaluated trials are stored in study.trials and we can access a
particular trial by its index. For example, above output tells us that
the first trial is failed. Check its state by
study.trials[0].state
TrialState.FAIL
According to the log output, the trial fails because the objective
value happens to be NaN. Please see the doc for more explanations on
trial states in optuna. Actually, the instances stored in study.trials
are instances of FrozenTrial, and contain the status and results of
existed trials; see the doc for more details on the methods and
attributes of frozen trials.
The optimize method can be called multiple times and the study object
would preserve the entire optimization history. For instance, here we
run another 20 trials, which are numbered starting from 10.
study.optimize(objective, n_trials=20)
The study object provides useful interfaces to inspect the experiment
results. For example, the trials_dataframe method can export all trial
results as a pandas dataframe, and the best_trial attribute returns
the trial result with best objective value. See the API reference for
more details.
Save and visualize
We can save the study object to a RDB (Relational DataBase) for future
usage. In general, all data associated with a Study instance is stored
in its attribute _storage, which is created at initialization; see the
API reference for details. As we did not manually specify the storage,
optuna initializes in-memory storage by default. In addition, optuna
also automatically assigns a name to the study if not specified.
study.study_name, study._storage
('no-name-1303842b-a35f-4d3e-b74f-15b25601b7ff',
<optuna.storages._in_memory.InMemoryStorage at 0x7d9e495c5cf0>)
Optuna does not provide a direct method for saving an in-memory study to an RDB, at least I did not find it in the doc at the time of writing this post. Nevertheless, we can create a new study with RDB storage and copy the in-memory study to the new one.
# save by copying to a new study with RDB storage study_saved = optuna.copy_study( from_study_name=study.study_name, from_storage=study._storage, to_storage="sqlite:///fit-sin.db", to_study_name="Random Search", )
This will save all trial results in a sqlite database ./fit-sin.db;
see here for how to set up the database URL. See also this official
tutorial for how to work with RDB backend.
Like in-memory storage, RDB-based storage is continuously updated
during the optimization process, and we can view the real-time
progress with optuna-dashboard.
optuna-dashboard sqlite:///fit-sin.db
Listening on http://127.0.0.1:8080/
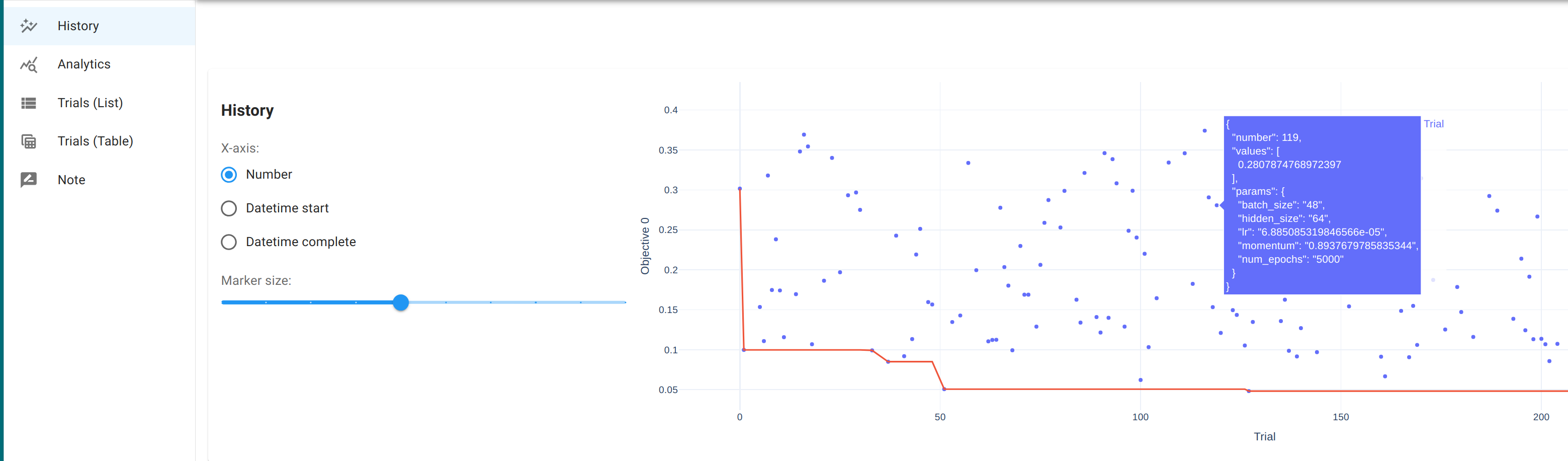
By default, this launches a local HTTP server at localhost:8080. Open
it in the browser to access the dashboard, which provides an
interactive interface for visualizing the optimization history. See
the dashboard doc for more details. See also the doc for
optuna.visualization module for how these visualizations are generated
and what they mean.
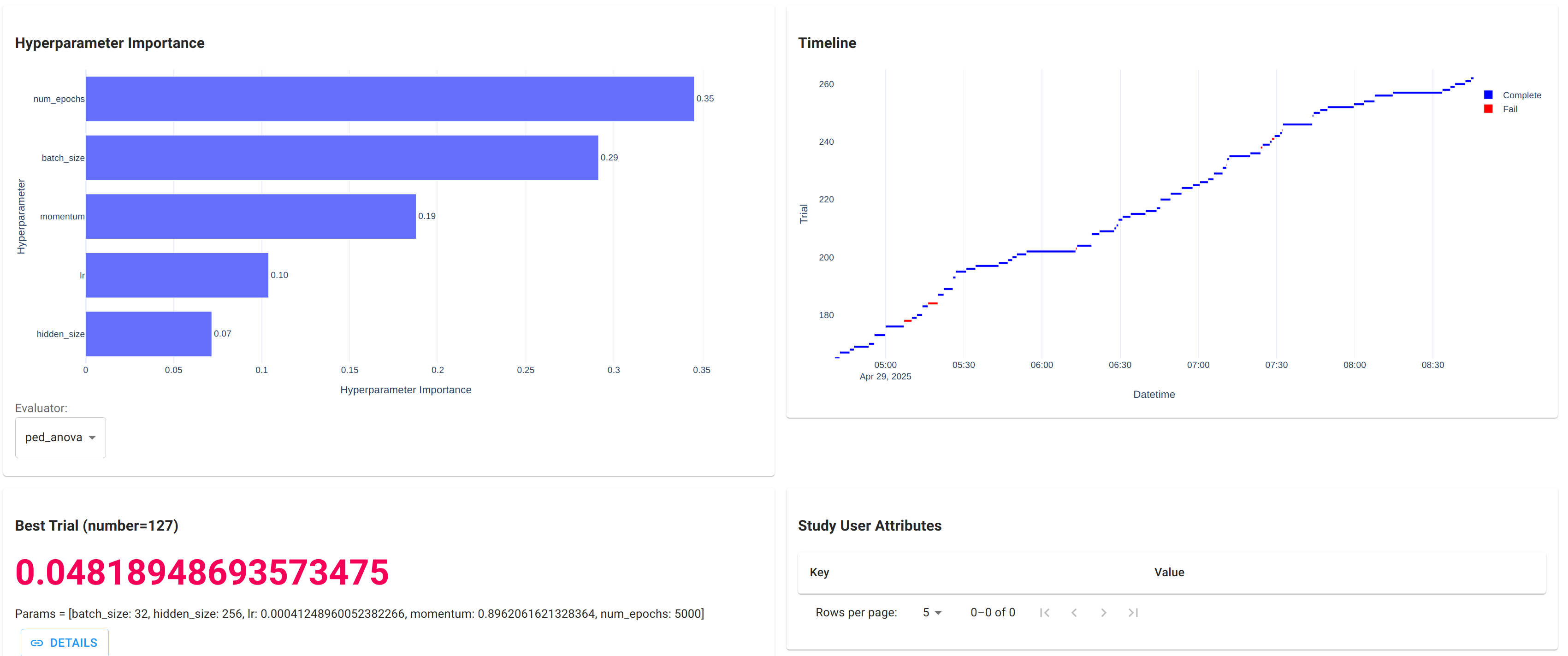
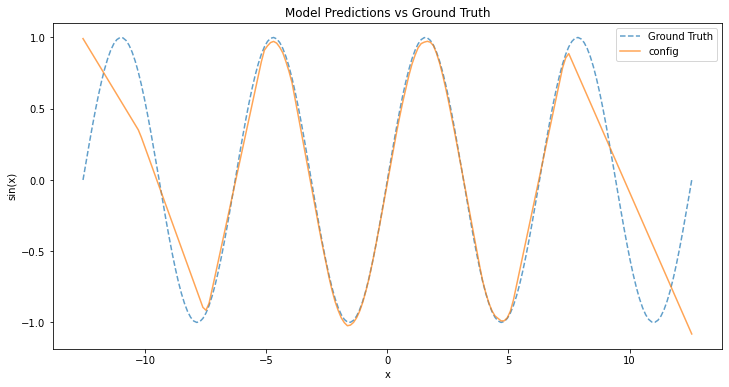
The best trial
In a following experiment, Optuna run a random search for 4 hours and explored 265 possible hyperparameter combinations. The best trial achieved objective value 0.048 with
batch_size 32 hidden_size 256 lr 0.00041248960052382266 momentum 0.8962061621328364 num_epochs 5000
Training the model with that combination yields the following function curve.
Advanced Features
The previous section covered the basic usage of Optuna. The next step is to integrate Optuna into various aspects of the training workflow.
- Explore the hyperparameter space. Random search is simple and efficient for most problems, but Optuna also provides many sophisticated samplers; see its manual for more discussion.
- Prune unpromising trials early. The pruner is another core concept of Optuna. By monitoring the test error during the training process, it decides whether to terminate the trial before completion and saves time for other trials.
- Search endlessly until manual termination. It's easy to write a
script and let Optuna do trials endlessly in a remote machine until
being manually terminated or reaching elapsed time; see the doc of
study.optimizefor more details. - Run multiple trials by parallelization. Parallelization and distributed calculation in Optuna are straightforward; see this official tutorial for an example.
- Manage trials results. Using Optuna's RDB API, it is straightforward to add or copy existing trial results. This enables use cases like saving and resuming the study via loading all results in the database; see also this official tutorial.
For more advanced features, please refer to the official tutorial collections.
References
- Optuna's official documentation
- Optuna. (2018). Optuna: A hyperparameter optimization framework. Read the Docs. https://optuna.readthedocs.io/en/stable/index.html
- Optuna's paper
- Akiba, T., Sano, S., Yanase, T., Ohta, T., & Koyama, M. (2019). Optuna: A next-generation hyperparameter optimization framework. Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 2623–2631. https://doi.org/10.1145/3292500.3330701
Appendix: Document the Experiment
It is good practice to document our findings and ideas during exploration. For instance, create a dedicated text file to serve as a lab journal. The actual format can vary based on requirements; the example below is just one possible approach.
In this journal, clearly state the problem and the expected
outcome. Whenever we explore a certain idea, create a dedicated entry
to elaborate the motivation and record findings. To simplify
referencing, name these entries using a consistent pattern, e.g.,
Study-DDDD-Title, where Title is a brief study description and DDDD is
a four-digit ID generated randomly. Each entry should include the
following elements:
- Date. Specify when the study was conducted.
- Idea. Describe the motivation and basic information about this study.
- Setup. Explain how the experiment was configured to ensure reproducibility.
- Result. Record the outcomes..
- Analysis. Summarize insights and interpretations.
- Other. Additional notes or observations.
In practice, we can use the Git commit hash for describing the experiment configuration and record results in a RDB via Optuna.
The journal I created for this problem is attached here.